Examples¶
Load the PMF¶
Given an output folder in /home/myname/Documents/PMF/GRE-cb/MobilAir_woOrga that looked like:
MobilAir_woOrga
├── GRE-cb_BaseErrorEstimationSummary.xlsx
├── GRE-cb_base.xlsx
├── GRE-cb_boot.xlsx
├── GRE-cb_ConstrainedDISPest.dat
├── GRE-cb_ConstrainedDISPres1.txt
├── GRE-cb_ConstrainedDISPres2.txt
├── GRE-cb_ConstrainedDISPres3.txt
├── GRE-cb_ConstrainedDISPres4.txt
├── GRE-cb_ConstrainedErrorEstimationSummary.xlsx
├── GRE-cb_Constrained.xlsx
├── GRE-cb_diagnostics.xlsx
├── GRE-cb_DISPest.dat
├── GRE-cb_DISPres1.txt
├── GRE-cb_DISPres2.txt
├── GRE-cb_DISPres3.txt
├── GRE-cb_DISPres4.txt
├── GRE-cb_Gcon_profile_boot.xlsx
├── GRE-cb_rotational_comments.txt
└── GRE-cb_sourcecontributions.xls
in order to convert them to a PMF object, run the following command :
from py4pm.pmfutilities import PMF
grecb = PMF(site="GRE-cb", BDIR="/home/myname/Documents/PMF/GRE-cb/MobilAir_woOrga")
Now, grecb is an instance of a PMF object, and has a lot of reader and plotter.
Read the data¶
Organization¶
The read class of the PMF object give access to different reader to retreive data from
the different xlsx files outputed by the EPA PMF5 software.
They all start by read_base* or read_constrained* name, for the base and constrained
run, respectively.
The special method read_metadata is used to retrieve the factors names and species names
from the _base.xlsx files, and use them everywhere else. It also try to set the total
variable name if any (one of PM10, PM2.5, PMrecons, PM10rec, PM10recons, otherwise try to
guess), used to convert unit and to be the default variable to plot.
For now, the following readers are implemented :
Contribution¶
The contributions of the factors (G matrix) are read from the _base.xlsx and
_Constrained.xlsx files, sheet contributions.
You can read them using the reader read_base_contributions and
read_constrained_contributions:
grecb.read.read_base_contributions()
grecb.read.read_constrained_contributions()
And now, the grecb object has a dfcontrib_b and dfcontrib_c attributes (_b for the
base run, _c for the constrained run):
>>> grecb.dfcontrib_c
Sulfate-rich Nitrate-rich ... Biomass burning Sea/road salt Mineral dust
Date ...
2017-02-28 0.321580 -0.105980 ... 0.19419 0.606290 0.182880
2017-03-03 0.429480 -0.038802 ... 0.61595 0.050129 0.382890
2017-03-06 -0.098123 -0.151530 ... 0.53346 4.636400 0.272410
2017-03-09 0.643500 -0.002527 ... 1.09060 0.153200 1.083600
2017-03-12 0.664090 0.308390 ... 1.70740 -0.200000 0.846930
which is the G matrix, in normalized unit.
Chemical profiles¶
The chemical profiles (or simply profiles) is the F matrix of the PMF (in µg/m³) and
are read from the _base.xslx and _Constrained.xlsx files, sheet Profiles.
You can read them using the reader read_base_profiles and read_constrained_profiles:
grecb.read.read_base_profile()
grecb.read.read_constrained_profile()
and grecb has now a not null dfprofiles_b and dfprofiles_c dataframe :
>>> grecb.dfprofiles_c
Sulfate-rich Nitrate-rich ... Biomass burning Sea/road salt Mineral dust
specie ...
PMrecons 4.402500 2.421300 ... 3.027900 0.364280 2.009600
OC* 1.225300 0.000000 ... 1.308900 0.041038 0.428110
EC 0.162970 0.000000 ... 0.347050 0.019199 0.030703
Cl- 0.000000 0.002425 ... 0.026819 0.109070 0.000000
NO3- 0.300660 1.702200 ... 0.093396 0.000000 0.000000
SO42- 0.977680 0.010441 ... 0.092800 0.032969 0.189890
... ... ... ... ... ... ...
The values are in µg/m³.
Uncertainties¶
Summary¶
You can also read the bootstrap and DISP results from the
_BaseErrorEstimationSummary.xlsx and _ConstrainedErrorEstimationSummary.xlsx files.
grecb.read.read_base_summary()
grecb.read.read_constrained_summary()
and now, you have access to df_uncertainties_summary_b and df_uncertainties_summary_c:
the summaries of the BS, DISP and BS-DISP uncertainties for each profiles and species.
>>> grecb.df_uncertainties_summary_c
Constrained base run BS 5th BS median BS 95th BS-DISP 5th BS-DISP average BS-DISP 95th DISP Min DISP average DISP Max
profile specie
Sulfate-rich PMrecons 4.402500 4.261867 4.511374 4.709612 NaN NaN NaN 3.788500 4.337850 4.887200
OC* 1.225300 0.822712 1.161025 1.702325 NaN NaN NaN 0.988480 1.211690 1.434900
EC 0.162970 0.051262 0.211147 0.436615 NaN NaN NaN 0.121070 0.213030 0.304990
Cl- 0.000000 0.000000 0.000000 0.000000 NaN NaN NaN 0.000000 0.006156 0.012311
NO3- 0.300660 0.000000 0.346984 0.563892 NaN NaN NaN 0.068862 0.260436 0.452010
... ... ... ... ... ... ... ... ... ... ...
Mineral dust Se 0.000008 0.000000 0.000012 0.000029 NaN NaN NaN 0.000000 0.000023 0.000046
Sn 0.000000 0.000000 0.000032 0.000154 NaN NaN NaN 0.000000 0.000069 0.000139
Ti 0.002545 0.001121 0.001750 0.002546 NaN NaN NaN 0.002881 0.003464 0.004047
V 0.000265 0.000063 0.000145 0.000249 NaN NaN NaN 0.000265 0.000278 0.000290
Zn 0.000218 0.000000 0.000030 0.001286 NaN NaN NaN 0.000000 0.000177 0.000354
All bootstrap profiles¶
If you want to retreive the individual bootstrap results, read from
_boot.xlsx and _Gcon_profile_boot.xlsx:
grecb.read.read_base_bootstrap()
grecb.read.read_constrained_bootstrap()
and now you have access to dfBS_profile_b and dfBS_profile_c, which are all the
bootstrap chemical profiles for the base and constrained run, respectively.
>>> grecb.dfBS_profile_c
Boot0 Boot1 Boot2 ... Boot97 Boot98 Boot100
specie profile ...
PMrecons Sulfate-rich 4.412330 2.259480 4.330630 ... 3.191810 4.041220 3.109190
Nitrate-rich 2.462740 2.254470 2.609910 ... 2.068200 2.349640 2.404520
Industrial 0.259120 0.289952 0.474484 ... 0.214298 0.206250 0.875102
Primary biogenic 0.579702 1.437820 0.633064 ... 1.290640 0.358833 0.296207
Primary traffic 1.862990 1.178150 1.711440 ... 1.171830 1.974060 1.678320
... ... ... ... ... ... ... ...
Zn Marine SOA 0.000826 0.002239 0.001256 ... 0.000265 0.000389 0.000436
Aged seasalt 0.000000 0.000000 0.000000 ... 0.000814 0.000304 0.001018
Biomass burning 0.002404 0.001699 0.002053 ... 0.002012 0.002270 0.001188
Sea/road salt 0.000625 0.000457 0.000848 ... 0.000234 0.000596 0.000187
Mineral dust 0.000000 0.000000 0.000000 ... 0.001355 0.000000 0.000000
as well as dfbootstrap_mapping_b and dfbootstrap_mapping_c, which are the
tables of the mapping between reference and BS factors:
>>> grecb.dfbootstrap_mapping_c
Sulfate-rich Nitrate-rich Industrial Primary biogenic Primary traffic Marine SOA Aged seasalt Biomass burning Sea/road salt Mineral dust unmapped
BF-Sulfate-rich 94 0 2 0 3 0 0 0 0 0 0
BF-Nitrate-rich 0 99 0 0 0 0 0 0 0 0 0
BF-Industrial 0 0 99 0 0 0 0 0 0 0 0
BF-Primary biogenic 0 0 0 99 0 0 0 0 0 0 0
BF-Primary traffic 0 0 0 0 99 0 0 0 0 0 0
BF-Marine SOA 0 0 0 0 1 98 0 0 0 0 0
BF-Aged seasalt 0 0 0 0 0 0 99 0 0 0 0
BF-Biomass burning 0 0 0 0 0 0 0 99 0 0 0
BF-Sea/road salt 0 0 0 0 0 0 0 0 99 0 0
BF-Mineral dust 0 0 0 0 0 0 0 0 0 99 0
Plot utilities¶
For now, the following plotters are implemented :
Chemical profile (per microgram of total variable)¶
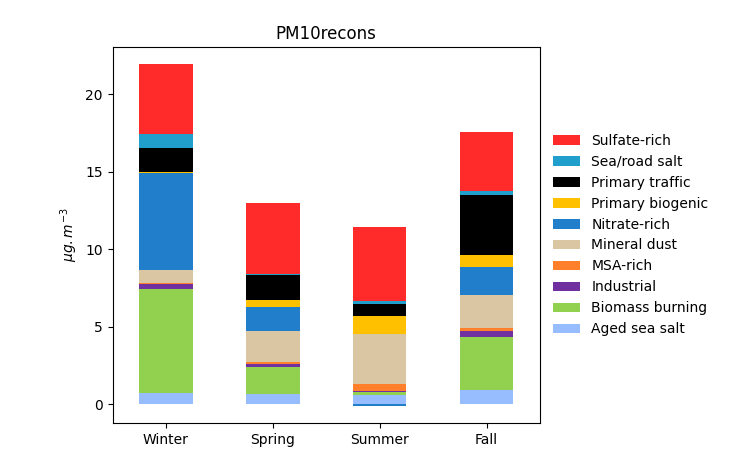
grecb.plot.plot_per_microgramm(profiles=["Primary biogenic"])
Chemical profile (in percentage of the sum of each species)¶
grecb.plot.plot_totalspeciesum(profiles=["Primary biogenic"])
Chemical profile stacked (in percentage of the sum of each species)¶
grecb.plot.plot_stacked_profiles()
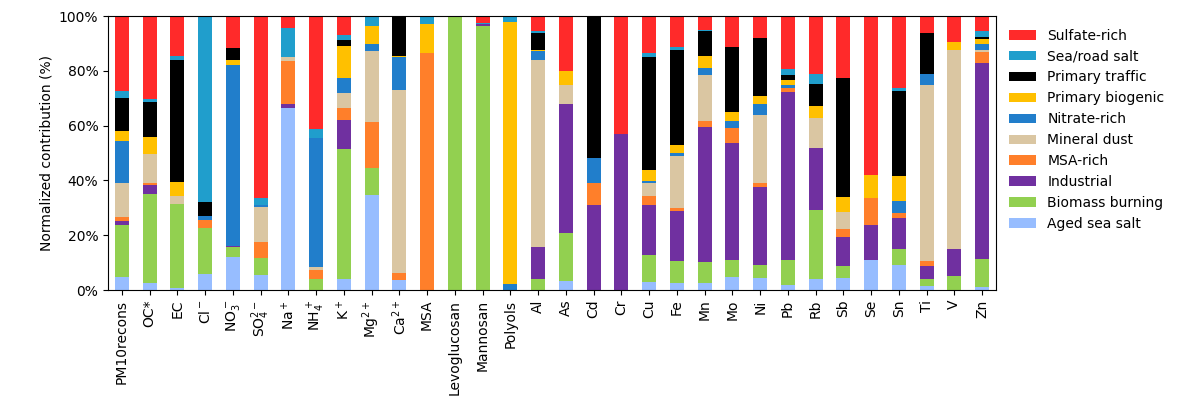
Contribution of each factor to the different species¶
Contribution time series and uncertainties¶
grecb.plot.plot_contrib(profiles=["Primary biogenic"])
will produce the following graph
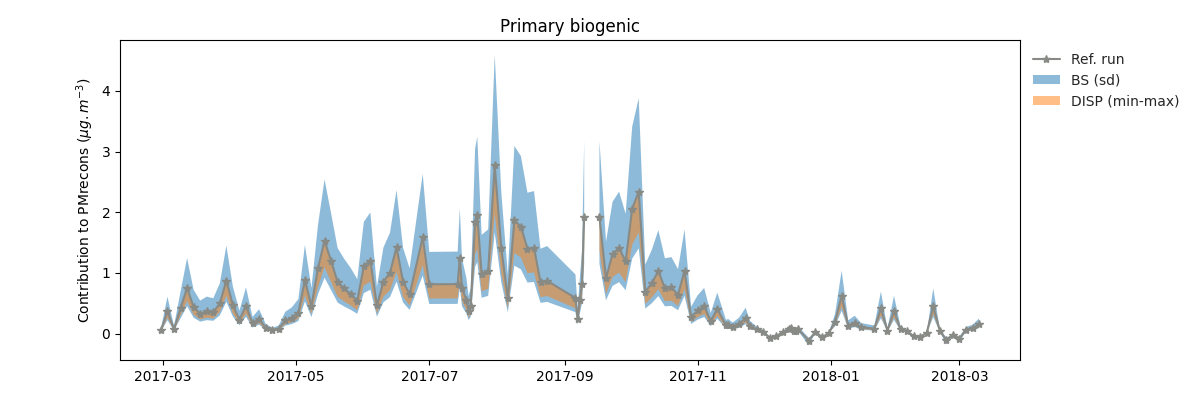
Primary biogenic factor contribution to the total variable.¶
Since the EPA PMF5 does not output the chemical profile (F) matrix of the boostrap, the uncertainties is estimated by computing the species concentration given the F matrix of the reference run and the G matrix of the bootstrap run. As a result, the output is “hacky” since in the bootstrap method, bith the F and G matrix are changing. If you want to remove them, just pass BS=False to the method.
Utilities¶
Convert to cubic meter¶
In order to have the contributions in µg/m³, which is given by G⋅F, we need to know
both the chemical profile F and the contribution G.
And we can easily reconstruct the time serie in µg/m³ of each specie for every profile
by simple multiplication of the timeserie by the concentration in the chemical profile.
Since this is a very often computation, the method to_cubic_metter does just that :
>>> grecb.to_cubic_metter()
Sulfate-rich Nitrate-rich ... Biomass burning Sea/road salt Mineral dust
Date ...
2017-02-28 1.415756 -0.256609 ... 0.587988 0.220859 0.367516
2017-03-03 1.890786 -0.093951 ... 1.865035 0.018261 0.769456
2017-03-06 -0.431987 -0.366900 ... 1.615264 1.688948 0.547435
2017-03-09 2.833009 -0.006120 ... 3.302228 0.055808 2.177603
2017-03-12 2.923656 0.746705 ... 5.169836 -0.072856 1.701991
... ... ... ... ... ... ...
Note that to_cubic_metter use by default the constrained run, all the profile
and the total variable, but you can specify other conditions (see the doc of
this method).
Relative contributions of species to the total mass¶
By default, the profile matrix F is in µg/m³. But it’s often convenient to know the
relative contribution of each species to the “total variable” mass (for instance, percent of
contribution of each specie to the $PM_10$).
This result is the ratio of each species in a profile to the total variable.
The method to_relative_mass conveniently handle it, and return you a new dataframe:
>>> grecb.to_relative_mass()
Sulfate-rich Nitrate-rich ... Biomass burning Sea/road salt Mineral dust
specie ...
PMrecons 1.000000 1.000000 ... 1.000000 1.000000 1.000000
OC* 0.278319 0.000000 ... 0.432280 0.112655 0.213032
EC 0.037018 0.000000 ... 0.114617 0.052704 0.015278
Cl- 0.000000 0.001002 ... 0.008857 0.299413 0.000000
NO3- 0.068293 0.703011 ... 0.030845 0.000000 0.000000
SO42- 0.222074 0.004312 ... 0.030648 0.090505 0.094491
... ... ... ... ... ... ...
The values are now in % of the PMrecons mass.
Relative contribution of the factor for each species¶
Another usefull information is how much a given specie is apportioned by all factors, denoted as the total specie sum graph in the EPA PMF5 software. It is the amount of a given specie in a factor divided by the sum of this specie in all factors.
The method get_total_specie_sum return this value for every species in all profiles:
>>> grecb.get_total_specie_sum()
Sulfate-rich Nitrate-rich ... Biomass burning Sea/road salt Mineral dust
specie ...
PMrecons 27.520080 15.135575 ... 18.927439 2.277119 12.562033
OC* 30.440474 0.000000 ... 32.517372 1.019519 10.635658
EC 14.525003 0.000000 ... 30.931475 1.711146 2.736462
Cl- 0.000000 1.506558 ... 16.659544 67.752580 0.000000
NO3- 11.676593 66.107550 ... 3.627177 0.000000 0.000000
SO42- 66.571611 0.710942 ... 6.318883 2.244906 12.929878
... ... ... ... ... ... ...
In this example, the Biomass burning factor apportion 18% of the total PMrecons, 32% of the OC*, 30% of the EC, etc. We also see that the NO3- is mainly apportioned by the Nitrate-rich factor (66%).